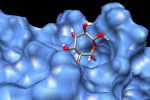
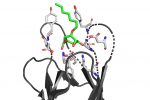
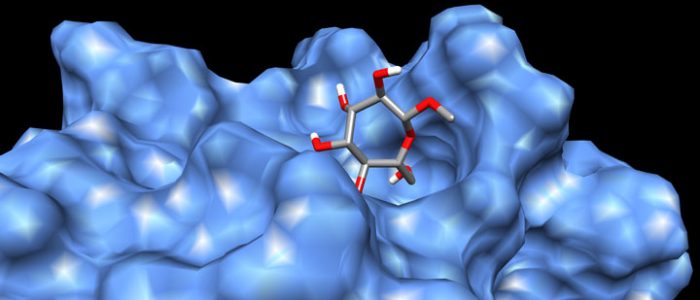
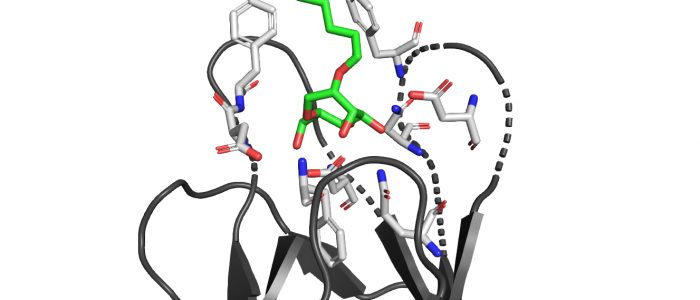
 QM/MM computed complex between ConA and a methyl septanoside.
QM/MM computed complex between ConA and a methyl septanoside.
 Cava is the only way to celebrate a successful thesis defense.
Cava is the only way to celebrate a successful thesis defense.

 @SammyKatta
@SammyKatta
About
The Peczuh Research Group investigates the inter-relationship between structure and function of small molecules in the context of biological activity. Organic synthesis is an integral tool in our research program. We develop new routes to molecules of intermediate complexity to address structure-activity relationships. The Group supports an inclusive research environment that encourages inquiry through rigorous experimentation, sound logic and unconventional thinking.
What We’re Reading
Journal: Science
Journal: Nature
Journal: ACS Publications
Journal: RSC Publications
Journal: Wiley Chemistry
Blog: In the Pipeline
Blog: Chemjobber
Blog: Master Organic Chemistry
Useful Links
The Offset
- Accumulating a foundationFull Title: “Accumulating” a foundation: The philosophy behind organic cumulative exams1 Michael Jordan, the greatest basketball player of all time, put it best when he said, “Get the fundamentals down and the level of everything you do will rise.” Cumulative exams are about setting the foundation for your training. They are a signature part of […]
- Twenty Year TakeawaysThe headline from the recent “Building UConn Chemistry for Another 20 Years,” panel discussion wasn’t the whiz-bang technical stuff you might expect from an event like this, although that element was definitely present. What really jumped out were the timeless bits of advice from the panelists: “Challenge the conventional wisdom.” – Gary Brudvig, Yale University […]
- Neighboring Group ParticipationFor Toni Planas, a sabbatical in rural Connecticut has accelerated a sweet collaboration with Mark Peczuh’s research group. The Glycosidase Team: (L to R) Sergi Pascual, Aditya Pote, Antoni Planas, Mark Peczuh Carbohydrate chemists are readily familiar with the concept of neighboring group participation (NPG), where the electrons of…
- Fischer Fool-Proof: Lessons from Stereochemistry’s Shining DebutAn explication of Fischer’s proof for the configuration of sugars is the capstone lecture in sophomore organic chemistry. This feat of experimentation and logic offers lessons that are of broad utility beyond organic chemistry.
- Hypocrellin A, With a TwistSome time ago my research group stumbled upon a family of macrocycles whose structure contained a somewhat uncommon stereogenic element, a plane of chirality. A plane of chirality arises when restricted rotation around a planar unit in a molecule leads to the formation of stereoisomers. The textbook example (seriously, look at Eliel) is E-cyclooctene. The […]
- Still Working Toward Convergence in Total SynthesisI was at the Natural Products and Bioactive Compounds Gordon Research Conference (GRC) last summer. During the usual free time one afternoon, there was a roundtable discussion about the challenges faced by women in science and strategies to overcome those challenges. The conversation was promoted by the GRC organization, which made sure there was an […]
We took our revenge on carbohydrates by attacking some cellulosic materials with axes yesterday at Pine and Iron Axe Throwing in #Hartford.
Thx @todd_hyster and @pfizer scientists for talking w/ @peczuh_research at the @ACS_CVS poster session yesterday. @swimmerkarate
@peczuh_research standing tall at today’s @ACS_CVS organic symposium. cc: @swimmerkarate
Summer Squad 2019 (minus @swimmerkarate): Jean, Bryant, Matthew, @mwpeczuh, Caleb, Yotam. We miss you CC! Come back from your @Boehringer internship already. #REU #UConnChem